mRNA Degradation
A recent study has generated some excitement and concern
In short it states the following
“The observed extended presence of vaccine mRNA and spike protein in vaccinee LN GCs for up to 2 months after vaccination was in contrast to rare foci of viral spike protein in COVID-19 patient LNs.
LN=Lymph Nodes
Spike protein was detected in the plasma of 96% of the vaccinees at days 1-2 (median spike concentration of 47 pg/mL) and in 63% at day 7 (median spike concentration of 1.7 pg/mL) after. the
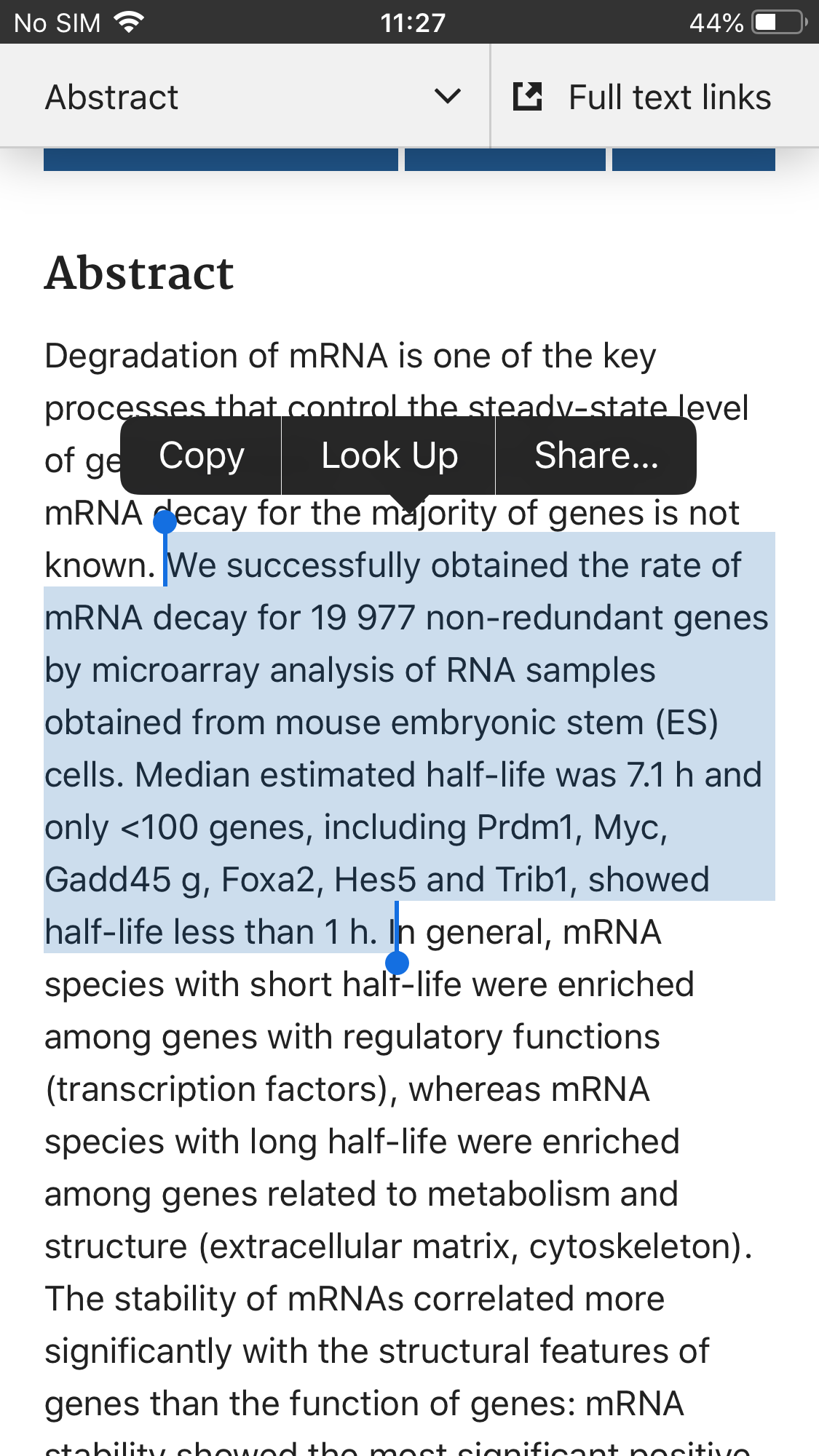
At least some portion of spike antigen generated after administration of BNT162b2 becomes distributed into the blood. We detected spike antigen in 96% of vaccinees in plasma collected one to two days after the prime injection, with antigen levels reaching as high as 174 pg/mL.
175 pg/ml comes out to 0.5 micrograms in a typical adult. The Pfizer dose of mRNA is 30 micrograms. So its likely this is just a small fraction of what is produced is in the blood plasma
In fact according to my ball park estimate (+/- order of magnitude ] at 100% transfection/translation efficiency efficiency about 4300 micrograms of spike is expected (most of it in injection site but we dont know cause Pfizer wont tell us

Now it helps to remember what we were told by FDA
“The delivered mRNA does not enter the cell nucleus or interact with the genome, is nonreplicating, and is expressed transiently. The estimated half-life for mRNA after injection is approximately 8 to 10 hours, before degradation by native RNases in the body, but the duration of effect also depends on the half-life of the expressed protein, which persists in the body for several days. mRNA vaccines have been used to induce immune responses against infectious pathogens such as cytomegalovirus (CMV), human metapneumovirus (hMPV) and parainfluenza virus type 3 (PIV3), Zika, and influenza virus.”
Pg 18
https://www.fda.gov/media/144452/download
Lets see what else is in this exciting paper. My comments in brackets
“mRNA vaccination was associated with follicular hyperplasia with fully developed GC architecture, including robust induction of GC B cells, Tfh cells and extensive follicular dendritic cell networks (Figures 6A and 6B).
The biodistribution, quantity and persistence of vaccine mRNA and spike antigen after vaccination, and viral antigens after SARS-CoV-2 infection, are incompletely understoodbut are likely to be major determinants of immune responses.
[They are incompletely understood be because FDA did not do its job and require this to be done in animals. This could have been done in parallel with human trials]
We performed in situ hybridization with control and SARS-CoV-2 vaccine mRNA-specific RNAScope probes
in the core needle biopsies of the ipsilateral axillary LNs that were collected 7-60 days after 2nd dose of mRNA-1273 or
BNT162b2 vaccination, and detected vaccine mRNA collected in the GCs of LNs on day 7, 16, and 37 post vaccination, with lower but still appreciable specific signal at day 60
[but is the mRNA Transcript competent or is it degraded. Could this be defective or truncated mRNA which did not produce protein so did not undergo degradation ]
Only rare foci of vaccine mRNA were seen outside of GCs.
Axillary LN core needle biopsie of non-vaccinees (n = 3) and COVID-19 patient specimens were negative for vaccine probe hybridization.
Immunohistochemical staining for spike antigen in mRNA vaccinated patient LNs varied between individuals, but showed abundant spike protein in GCs 16 days post-2nd dose, with spike antigen still present as late as 60 days post-2nd dose.
Spike antigen localized in a reticular pattern around the GC cells, similar to staining for follicular dendritic cell processes COVID-19 patient LNs showed lower quantities of spike antigen, but a rare GC had positive staining .
Spike protein was detected in the plasma of 96% of the vaccinees at days 1-2 (median spike concentration of 47 pg/mL) and in 63% at day 7 (median spike concentration of 1.7 pg/mL) afterthe prime vaccine dose.
The observed extended presence of vaccine mRNA and spike protein in vaccinee LN GCs for up to 2 months after vaccination was in contrast to rare foci of viral spike protein in COVID-19 patient LNs.
Persistent vaccine RNA and spike antigen at elevated concentrations in vaccinee LNs could result in less strict selection for higher-affinity B cells in the immune response compared to situations where antigen is more limiting (Cirelli et al., 2019).
Pre-pandemic analysis of a model RNA vaccine for yellow fever virus in a rhesus macaque at 16 hours post-vaccination showed that vaccine RNA in LN cell suspensions was detected predominantly in professional antigen-presenting cells including monocytes, classical dendritic cells and B cells at this early time point (Lindsay et al., 2019). Data from follicular dendritic cells were not reported.
[I am not an immunologist but transfecting immune cells with spike producing mRNA cant do your immune system any good. Perhaps this explains the transient (?) immune suppression some are reporting]
Our histological data from SARS-CoV-2 mRNA-vaccinated humans at considerably later time points (7 to 60 days post-2nd dose) show vaccine RNA almost entirely in GC’s , distributed primarily between the nuclei of GC cells, similar to the pattern seen by immunostaining for follicular dendritic cell processes or B cell cytoplasm.
[Are they saying the RNA was hanging out in the cell nucleus where its not supposed to be? Or am I misunderstanding this. Might explain why it escaped the house cleaning in cytoplasm]
Additional co-localization studies with higher resolution may be required to determine more exactly which specific cell types harbor mRNA vaccine and spike antigen in humans following COVID-19 mRNA vaccination and infection, and may provide further mechanistic insights into the basis for the differences in serological responses after vaccination compared to infection.
[Duh, maybe its hidden in the Pfizer documents they are so concerned about]
At least some portion of spike antigen generated after administration of BNT162b2 becomes distributed into the blood. We detected spike antigen in 96% of vaccinees in plasma collected one to two days after the prime injection, with antigen levels reaching as high as 174 pg/mL.
The range of spike antigen concentrations in the blood of vaccinees at this early time point largely overlaps with the range of spike antigen concentrations reported in plasma in a study of acute infection (Ogata et al., 2020), although a small number of infected individuals had higher concentrations in the ng/mL range.
[they looked at those with serious COVID. This is unlikely for those with mild or asymptomatic COVID]
At later time points after vaccination, the concentrations of spike antigen in blood quickly decrease, although spike is still detectable in plasma in 63% of vaccinees one week after the first dose.
A practical finding in our study is that the detection of spike antigen in plasma samples is impeded after 2nd dose BNT162b2 vaccination, likely due to the formation of circulating immune complexes of anti-spike antibodies and spike protein, masking the antigen epitopes of the capture and detection antibodies that form the basis of antigen detection assays, similar to assay interference that has been reported for other diseases (Bollinger et al., 1992; Lima et al., 2014; Miles et al., 1993).
LN histological comparisons between COVID-19 patients and vacinees have the limitations that the infected patient specimens were limited to those with severe disease; the number of individuals analyzed was relatively low (six COVID-19 patients and seven vaccinees); and the LN sampling was not done prospectively at pre-determined time points aftervaccination or infection
[ok, that was interesting. We already knew the spike ends up in the plasma, and persistence in the GC is not all that surprising. However, the presence of mRNA in the GC after 60 days is surprising. Its not clear this is functional mRNA, or if its just fragments or complete mRNA. Unless Pfizer/Moderna have a quick explanation vaccinations should be halted.]
While we are at it lets look some more at mRNA degradation
This paper tells us
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8032477/
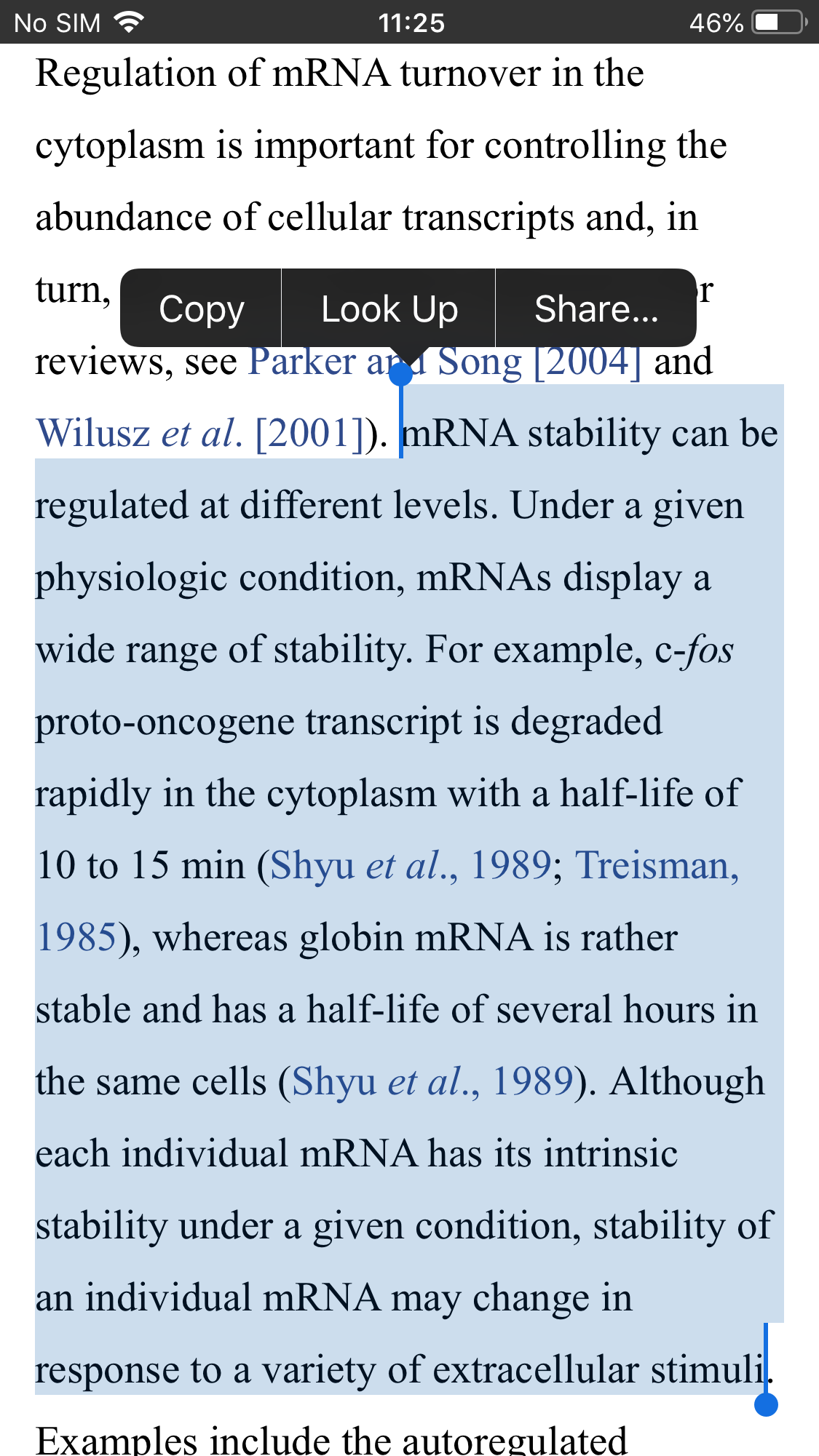
Many efforts have been made to increase the in vivo stability and translation capacity of the mRNA molecule, while avoiding unwanted innate immune activation.
One prevalent idea is that this can be achieved by optimizing the regulatory regions of the mRNA: the 5′ cap, poly-A tail and untranslated regions (UTRs). The UTRs are parts of the mRNA that flank the coding region of the mRNA and regulate its stability and translation.
The poly-A tail also regulates stability, as its shortening and eventual removal leads to mRNA degradation.
The 5′ cap structure is important for protein production and the recruitment of translation initiation factors (Pardi et al., 2018). Furthermore, mRNA with maximized GC (guanine-cytosine) content in combination with codon optimization, i.e., selection of ‘frequent codons’ in the coding region, leads to enhanced stability and translation (Thess et al., 2015).
Another critical determinant is the secondary structure of mRNA, which can be stabilized by changing the primary sequence through codon optimizations and computational tools. Building secondary structures in the mRNA –except in the 5′ UTR region– by choosing ‘highly structured coding sequences’ leads to higher translation in vivo as well, because of a longer functional half-life (Mauger et al., 2019).
Alternatively, Mauger et al. demonstrated that the incorporation of naturally occurring modified uridines, such as the use of 1-methyl-pseudouridine (m1Ψ) instead of uridine, induces global changes in the secondary structure of mRNA which correlated with high protein expression.
Importantly, the introduction of these modified uridines inside the mRNA construct is currently the most efficient way to minimize the cellular recognition of mRNA by RNA-binding proteins that are involved in the innate immune reaction to foreign mRNA.
This, in turn, enhances biological stability and translational capacity, while reducing the reactogenicity of mRNA vaccines. Moreover, there are also indications that m1Ψ increases base stacking and the melting point of mRNA, thereby making mRNA more stable (Mauger et al., 2019, Zhao and He, 2015).
This could mean that the incorporation of 1mΨ, as is done for the COVID-19 vaccines from Moderna and BioNTech/Pfizer, also affects the stability of mRNA before administration.
Another paper here
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2778729/#!po=0.568182
And here
https://pubmed.ncbi.nlm.nih.gov/19001483/
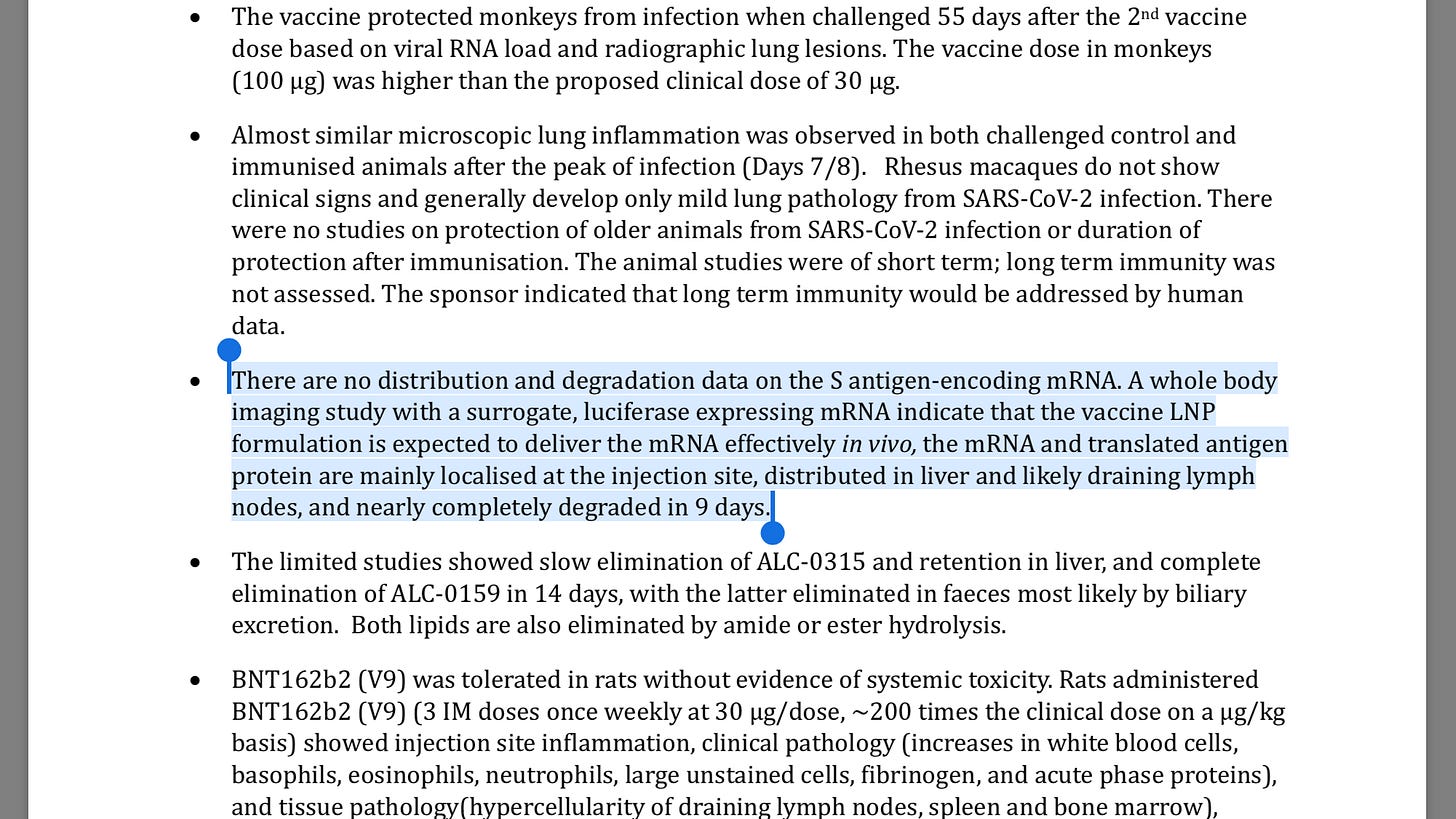
And here are Australia and EMA confirming no distribution and degradation studies done or required. Unimaginable. More likely is they did the studies but chose to hide the results.